Projects
publications
Check out my google scholar or ORCID for a list of my publications!
new projects ?
I am working in Dr. Jean Fan’s lab doing Spatial Transcriptomics analysis, so I look forward to what the future has in store!
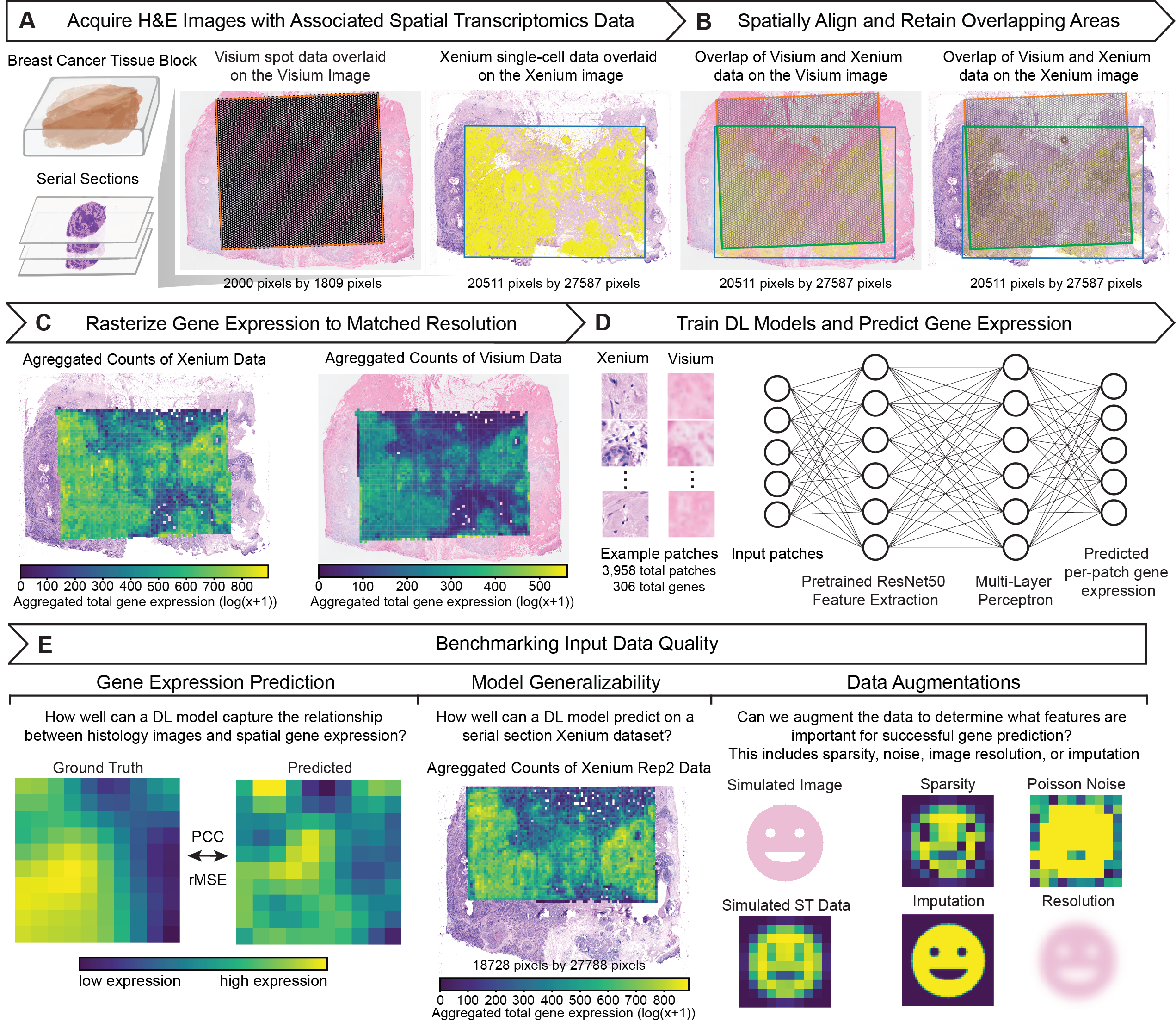
Impact of Data Quality on Deep Learning Prediction of Spatial Transcriptomics from Histology Images (2025)
Abstract
Spatial transcriptomics technologies enable high-throughput quantification of gene expression at specific locations across tissue sections, facilitating insights into the spatial organization of biological processes. However, high costs associated with these technologies have motivated the development of deep learning methods to predict spatial gene expression from inexpensive hematoxylin and eosin-stained histology images. While most efforts have focused on modifying model architectures to boost predictive performance, the influence of training data quality remains largely unexplored. Here, we investigate how variation in molecular and image data quality stemming from differences in imaging (Xenium) versus sequencing (Visium) spatial transcriptomics technologies impact deep learning-based gene expression prediction from histology images. To delineate the aspects of data quality that impact predictive performance, we conducted in silico ablation experiments, which showed that increased sparsity and noise in molecular data degraded predictive performance, while in silico rescue experiments via imputation provided only limited improvements that failed to generalize beyond the test set. Likewise, reduced image resolution can degrade predictive performance and further impacts model interpretability. Overall, our results underscore how improving data quality offers an orthogonal strategy to tuning model architecture in enhancing predictive modeling using spatial transcriptomics and emphasize the need for careful consideration of technological limitations that directly impact data quality when developing predictive methodologies.
Check out the bioRxiv preprint here.
Evidence of off-target probe binding in the 10x Genomics Xenium v1 Human Breast Gene Expression Panel compromises accuracy of spatial transcriptomic profiling (2025)
Abstract
The accuracy of spatial gene expression profiles generated by probe-based in situ spatially-resolved transcriptomic technologies depends on the specificity with which probes bind to their intended target gene. Off-target binding, defined as a probe binding to something other than the target gene, can distort a gene’s true expression profile, making probe specificity essential for reliable transcriptomics. Here, we investigate off-target binding in the 10x Genomics Xenium v1 Human Breast Gene Expression Panel. We developed a software tool, Off-target Probe Tracker (OPT), to identify putative off-target binding via alignment of probe sequences and found at least 21 out of the 280 genes in the panel impacted by off-target binding to protein-coding genes. To substantiate our predictions, we leveraged a previously published Xenium breast cancer dataset generated using this gene panel and compared results to orthogonal spatial and single-cell transcriptomic profiles from Visium CytAssist and 3’ single-cell RNA-seq derived from the same tumor block. Our findings indicate that for some genes, the expression patterns detected by Xenium demonstrably reflect the aggregate expression of the target and predicted off-target genes based on Visium and single-cell RNA-seq rather than the target gene alone. Overall, this work enhances the biological interpretability of spatial transcriptomics data and improves reproducibility in spatial transcriptomics research.
Check out the reviewed preprint on eLife here.
subtype discovery method utilizing scRNA-seq and microarray data (2025)
Our method, PHet, is able to distinguish multiple subtypes of data given only two labels (control and case)

Summary of Abstract
In disease diagnosis and targeted therapy, discovering subtypes is crucial as cells or patients can exhibit varied responses to treatments. Hence, understanding the heterogeneity of disease states is vital for comprehending pathological processes. However, selecting features for subtyping from high-dimensional datasets is challenging, with many algorithms focusing on known disease phenotypes and potentially overlooking valuable subtyping information. Our study aimed to address this issue by identifying feature sets that preserve heterogeneity while discriminating known disease states. Through a data-driven approach combining feature clustering and deep metric learning, we developed a statistical method called PHet (Preserving Heterogeneity). This method effectively identifies a minimal set of features that maintain heterogeneity while maximizing the quality of subtype clustering. PHet outperformed previous methods in identifying disease subtypes using microarray and single-cell RNA-seq datasets. Our research provides an innovative feature selection method that facilitates personalized medicine and enhances understanding of disease heterogeneity. I am co-first author with my former labmate Dr. Abdurrahman Abul-Basher.
Check out the publication in Nature Communications here.
image analysis of live-cell imaging (2022-2023)
I worked under Dr. Kwonmoo Lee at Boston Children’s Hospital as a Research Assistant for two years after undergrad. My work included using various Convolutional Neural Networks (CNNs) for cell segmentation, utilizing cell tracking algorithms, honing my image manipulation skills (Fiji), working on my research writing aptitude, and much more! It was an incredible experience that has led me to where I am now :) Below are two papers I had the privilege of working on:
The Lee Lab developed a deep learning-based pipeline termed MARS-Net . While I did not contribute to it’s development, I helped the first author write the protocol for running it here.
I helped in optimizing the hyper parameters for the R-CNN in FNA-Net, a deep-learning based ensemble model aimed to screen the adequacy of unstained thyroid fine needle aspirations (FNA). Ideally, this will streamline the diagnostic process by eliminating the need for staining and expert interpretation.
My biggest project was a subtype discovery method termed PHet. The abstract and preprint are above!
undergraduate research at the University of Virginia (2018-2021)
I started my research journey by collaborating with two incredible mentors, Dr. Tianxi Li and Dr. Frederic Padilla. Under Dr. Li’s guidance, I honed my coding skills in Python and R by converting his randnet package from R to Python. Later, I teamed up with Dr. Padilla to explore the effects of focused ultrasound on mouse tumors, which involved applying machine learning algorithms and statistical tests to understand its impact. In my final project my fourth year, I focused on classifying brain tumor regions in contrast-enhanced ultrasound images using microbubble intensity. I presented my final work via zoom (bc covid) where I expressed how well various unsupervised and supervised methods faired in this task. Though I didn’t publish any papers or achieve significant outcomes, this experience was incredibly valuable and marked the beginning of my research journey. I am immensely grateful to both mentors for the opportunity they gave me, as I don’t think I would be where I am now without them!